Smoother - A unified spatial dependency framework in PyTorch
Smoother is a Python package built for modeling spatial dependency and enforcing spatial coherence in spatial omics data analysis. Implemented in Pytorch, Smoother is modular and ultra-efficient, often capable of analyzing samples tens of thousands of spots in seconds.
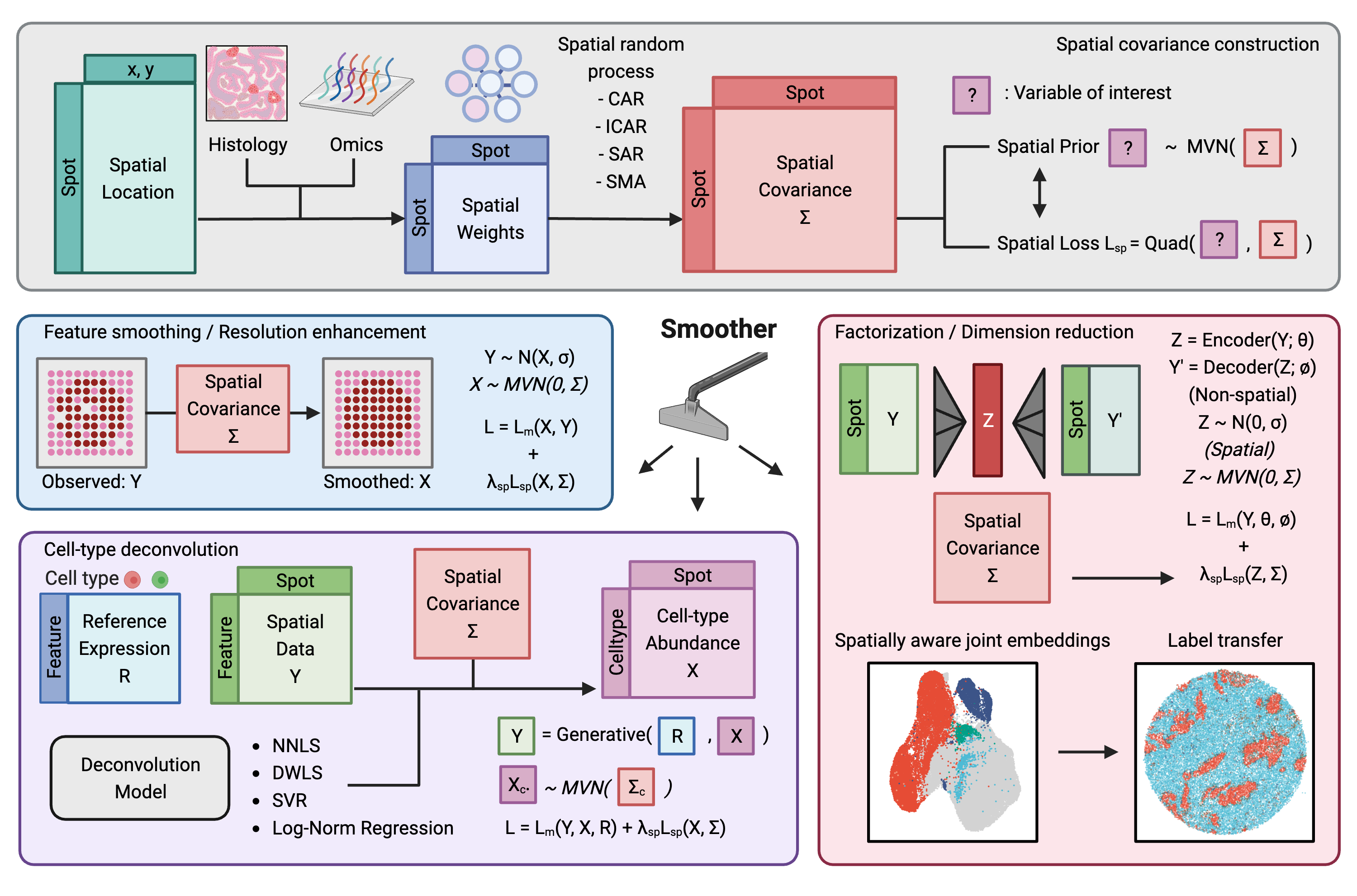
The key innovation of Smoother is the decoupling of the prior belief on spatial structure (i.e., neighboring spots tend to be more similar) from the likelihood of a non-spatial data-generating model. This flexibility allows the same prior to be used in different models, and the same model to accommodate data with varying or even zero spatial structures. In other words, Smoother can be seamlessly integrated into existing non-spatial models and pipelines (e.g. single-cell analyses) and make them spatially aware. In particular, Smoother provides the following functionalities:
Spatial loss: A quadratic loss equivalent to a multivariate Gaussian (MVN) prior reflecting the spatial structure of the data. It can be used to regularize any spatial random variable of interest.
Data imputation: Mitigates technical noise by borrowing information from the neighboring spots. It can also be applied to enhance the resolution of the data to an arbitrary level in seconds.
Cell-type deconvolution: Infers the spatially coherent cell-type composition of each spot using reference cell-type expression profiles. Smoother is one of the few deconvolution methods that actually enforce spatial coherence by design.
Dimension reduction: Find the spatially aware latent representations of spatial omics data in a model-agnostic manner, such that single-cell data without spatial structure can be jointly analyzed using the same pipeline.
For method details, check the Smoother paper (Su Jiayu, et al. 2023) and the Supplementary Notes. Check out the Installation and Basic usage sections for further information, including installation instructions and example usages.
Note
This documentation is under active development.
Citation
Su, Jiayu, et al. “Smoother: a unified and modular framework for incorporating structural dependency in spatial omics data.” Genome Biology 24.1 (2023): 291. https://link.springer.com/article/10.1186/s13059-023-03138-x